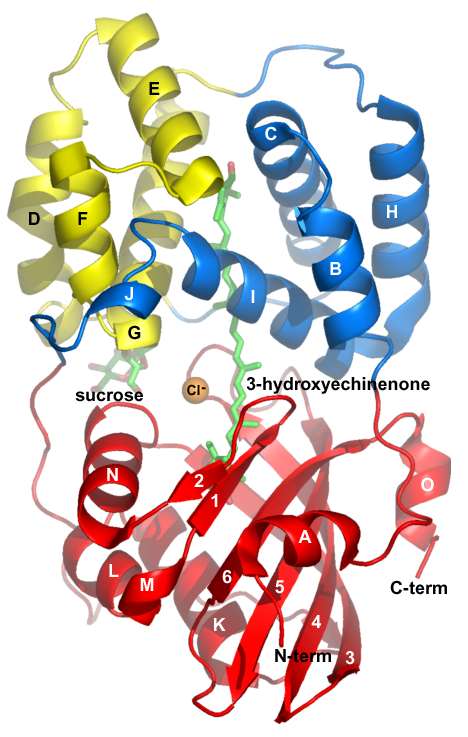
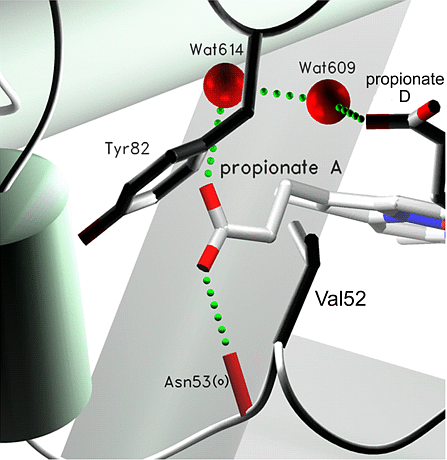
|
|
|
|
|
|
Pymol
Pros: 1) Ray tracing gives a real 3D quality. Background fog adds to the 3D effect and simplifies figures by hiding less important features in the background. 2) Can display molecular surfaces on the fly. 3) Runs on PC or linux machines. But, faster performance on linux. 4) Hardware stereo capability. 5) Displays X-plor format electron density maps. 6) Can display unit cell or dashed lines between atoms. 7) Can make movies in which your molecule rocks about the y axis. 8) Can show crystal packing. 9) If you write a pymol script, you can reproduce a figure by executing the script. Cons: 1) Menus are clumsy and incomplete. You need to learn pymol syntax to get anywhere. 2) Doesnt write out a pymol script. You write it yourself. 3) No built in capability to draw C-Alpha traces, although it can show backbone trace. References: See Pymol web page - for links to manual, gallery, etc. |
|
|
Ribbons 1) Beautiful ribbons display; ribbon style can be interactively customized. 2) Can display masks or molecular envelops with nice shadow effect. 3) Makes nice figures of DNA too. 4) Works on a DEC Alpha station, (but some features only work on SGI machines). 5) Quick interactive control over Orientation and ribbon dimensions. Cons: References: M. Carson (1997) Ribbons. Methods in Enzymology 277:493-505, R.M. Sweet and C.W. Carter, eds, Academic Press |
|
|
Molscript/Raster3D 1) Beautiful ribbons display. 2) Nice shadow effect. 3) Works on a DEC Alpha station, (Linux version not yet installed). 4) Scripted. Cons: References: Merritt, Ethan A. and Bacon, David J. (1997). "Raster3D: Photorealistic Molecular Graphics"277, 505-524. Methods in Enzymology |
| SETOR |
SETOR (pronounced see-tor) Pros: Cons: References: SETOR: Hardware-lighted three-dimensional solid model representations of macromolecules. S.V. Evans. J Mol Graph (1993) Jun;11(2):134-8, 127-8 evans@flint.biochm.uottawa.ca |
|
|
References: |