|
|
|
|
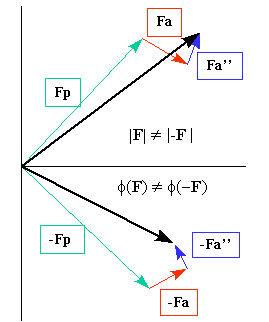
the contribution of anomalous scattering to the phasing calculation. Taken from a figure from Crystallography 101 |
MIRAS Phasing:A
sample
scriptis provided below for a typical MIRAS phasing calculation. All
steps on the path are provided from preparing the Scalepack files to displaying
the map with O. The general path is:
scalepack2mtz ->
Example log files are given for each step, along with comments about how to judge the quality of the output. Documentation for CCP4 programs:
References for CCP4 programs:
|
|
|
|
|
|
| #!/bin/csh -f
# scalepack2mtz hklin mtz/native.sca \ hklout mtz/native_i.mtz <<eof SYMM 90 ANOMALOUS NO END eof # scalepack2mtz hklin mtz/hg.sca \ hklout mtz/hg_i.mtz <<eof SYMM 90 ANOMALOUS YES END eof # scalepack2mtz hklin mtz/pt.sca \ hklout mtz/pt_i.mtz <<eof SYMM 90 ANOMALOUS YES END eof # scalepack2mtz hklin mtz/pb.sca \ hklout mtz/pb_i.mtz <<eof SYMM 90 ANOMALOUS YES END eof # |
CCP4 document for
scalepack2mtz
The native scalepack data set, native.sca, is converted to mtz format first, followed by three derivatives, Hg, Pt, and Pb. SYMM -This keyword is compulsory and can be given as the space group name or number. Here we are using space group number 90, otherwise known as P4212. ANOMALOUS -Specify whether or not input file contains anomalous data. It is set to NO for the native, and YES for the derivatives. ---------Logfile from scalepack2mtz----------
* Number of Columns = 9
|
|
|
|
|
|
| #!/bin/csh
-f
# truncate HKLIN mtz/native_i.mtz \ HKLOUT mtz/native_f.mtz \ << EOF_truncate > log/truncate_native.log TITLE native TRUNCATE OUTPUT LABOUT F=Fnat sigF=sigFnat NRESIDUES 410 EOF_truncate # truncate HKLIN mtz/hg_i.mtz \ HKLOUT mtz/hg_f.mtz \ << EOF_truncate > log/truncate_hg.log TITLE Hg TRUNCATE OUTPUT LABOUT F=Fhg sigF=sigFhg DANO=Dhg SIGDANO=sigDhg NRESIDUES 410 EOF_truncate # truncate HKLIN mtz/pt_i.mtz \ HKLOUT mtz/pt_f.mtz \ << EOF_truncate > log/truncate_pt.log TITLE Pt TRUNCATE OUTPUT LABOUT F=Fpt sigF=sigFpt DANO=Dpt SIGDANO=sigDpt NRESIDUES 410 EOF_truncate # truncate HKLIN mtz/pb_i.mtz \ HKLOUT mtz/pb_f.mtz \ << EOF_truncate > log/truncate_pb.log TITLE Pb TRUNCATE OUTPUT LABOUT F=Fpb sigF=sigFpb DANO=Dpb SIGDANO=sigDpb NRESIDUES 410 EOF_truncate # |
CCP4 document for
truncate
The native and derivative data sets are processed with truncate. Truncate converts the intensities into structure factors by taking the square root. Hence, the input files have the extension _i.mtz and the output files have the extension _f.mtz. It also puts the data sets on an absolute scale. This makes it convenient for estimating heavy atom occupancies. LABOUT -Output labels for each column of data. These should be meaningful and easy to remember. NRESIDUES -The number of residues in the asymmetric unit. Used to put the data sets on an absolute scale. ---------Logfile from truncate----------
Truncate outputs some useful statistics. The Wilson B-factor is calculated and plotted: SCALE = exp( - intercept). Least squares straight line gives: B = 21.434 SCALE = 216.53 where F(absolute)**2 = SCALE*F(observed)**2*EXP(-B*2*SINTH**2/L**2) |
|
|
|
|
|
| #!/bin/csh
-f
# cad hklin1 mtz/native_f.mtz \ hklin2 mtz/hg_f.mtz \ hklin3 mtz/pt_f.mtz \ hklin4 mtz/pb_f.mtz \ hklout mtz/derivs_unscaled.mtz \ < CELL 93.127 93.127 114.335 90.000 90.000 90.000 RESOLUTION OVERALL 20.0 2.0 SYMMETRY 90 TITLE native=pbection, hgote, pt, pbection SCAL FILE 1 1.0 LABIN FILE 1 E1=Fnat E2=sigFnat CTYP FILE 1 E1=F E2=Q LABOUT FILE 1 E1=Fnat E2=sigFnat LABIN FILE 2 E1=Fhg E2=sigFhg E3=Dhg E4=sigDhg
LABIN FILE 3 E1=Fpt E2=sigFpt E3=Dpt E4=sigDpt
LABIN FILE 4 E1=Fpb E2=sigFpb E3=Dpb E4=sigDpb
|
CCP4 document for
truncate
The native and derivative data sets are processed with truncate. Truncate converts the intensities into structure factors by taking the square root. Hence, the input files have the extension _i.mtz and the output files have the extension _f.mtz. It also puts the data sets on an absolute scale. This makes it convenient for estimating heavy atom occupancies. LABOUT -Output labels for each column of data. These should be meaningful and easy to remember. NRESIDUES -The number of residues in the asymmetric unit. Used to put the data sets on an absolute scale. ---------Logfile from truncate----------
Truncate outputs some useful statistics. The Wilson B-factor is calculated and plotted: SCALE = exp( - intercept). Least squares straight line gives: B = 21.434 SCALE = 216.53 where F(absolute)**2 = SCALE*F(observed)**2*EXP(-B*2*SINTH**2/L**2) |
|
|
|
|
|
| #!/bin/csh
-f
# # scaleit hklin mtz/derivs_unscaled.mtz hklout mtz/derivs.mtz << eof >log/scaleit.log TITLE Scaling native and derivatives GRAPH H K L MODF REFINE ISOTROPIC LABI FP=Fnat SIGFP=sigFnat - FPH1=Fhg SIGFPH1=sigFhg - DPH1=Dhg SIGDPH1=sigDhg - FPH2=Fpt SIGFPH2=sigFpt - DPH2=Dpt SIGDPH2=sigDpt - FPH3=Fpb SIGFPH3=sigFpb - DPH3=Dpb SIGDPH3=sigDpb eof |
CCP4 document for
truncate
The native and derivative data sets are processed with truncate. Truncate converts the intensities into structure factors by taking the square root. Hence, the input files have the extension _i.mtz and the output files have the extension _f.mtz. It also puts the data sets on an absolute scale. This makes it convenient for estimating heavy atom occupancies. LABOUT -Output labels for each column of data. These should be meaningful and easy to remember. NRESIDUES -The number of residues in the asymmetric unit. Used to put the data sets on an absolute scale. ---------Logfile from truncate----------
Truncate outputs some useful statistics. The Wilson B-factor is calculated and plotted: SCALE = exp( - intercept). Least squares straight line gives: B = 21.434 SCALE = 216.53 where F(absolute)**2 = SCALE*F(observed)**2*EXP(-B*2*SINTH**2/L**2) |
|
|
|
|
|
| mlphare
HKLIN mtz/derivs.mtz\ HKLOUT mtz/derivs_ph.mtz < TITLE MAD phasing from acentric data, with four sites + three derivs. SYMM 90 ANGLE 10 THRESHOLD 2.5 0.5 PRINT AVF AVE RESO 20 2.0 EXCLUD SIGFP 3 CYCLES 20 LABI FP=Fnat SIGFP=sigFnat - FPH1=Fhg SIGFPH1=sigFhg DPH1=Dhg SIGDPH1=sigDhg - FPH2=Fpt SIGFPH2=sigFpt DPH2=Dpt SIGDPH2=sigDpt - FPH3=Fpb SIGFPH3=sigFpb DPH3=Dpb SIGDPH3=sigDpb LABO ALLIN PHIB=PHIO FOM=FOMO HLOUT HLA=HLA HLB=HLB HLC=HLC HLD=HLD EXCLUDE DISO 590 EXCLUDE DANO 200 RUN DERIV Se hg Lambda 0.971139 f' -3 f" 3.5 DCYCLE PHASE ALL REFCYC ALL KBOV ALL ATOM HG 0.386 0.273 0.091 1.000 1.000 BFAC 53.621 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM HG 0.474 0.253 0.114 1.000 1.000 BFAC 29.510 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM HG 0.485 0.255 0.373 1.000 1.000 BFAC 14.148 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM HG 0.217 0.068 0.394 1.000 1.000 BFAC 50.503 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM HG 0.381 0.241 0.115 1.000 1.000 BFAC 23.365 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM HG 0.256 0.088 0.372 1.000 1.000 BFAC 27.015 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 EXCLUDE DISO 430 EXCLUDE DANO 215 DERIV Se pt Lambda 0.978680 f' -5.5 f" 4.6 DCYCLE PHASE ALL REFCYC ALL KBOV ALL ATOM PT 0.736 0.114 0.113 1.000 1.000 BFAC 15.807 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM PT 0.250 0.578 0.627 1.000 1.000 BFAC 12.870 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 EXCLUDE DISO 370 EXCLUDE DANO 270 DERIV pb Lambda 0.978437 f' -8.5 f" 2.2 DCYCLE PHASE ALL REFCYC ALL KBOV ALL ATOM PB 0.221 0.444 0.047 1.000 1.000 BFAC 53.986 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM PB 0.366 0.373 0.199 1.000 1.000 BFAC 38.440 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM PB 0.112 0.076 0.285 1.000 1.000 BFAC 35.399 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 ATOM PB 0.121 0.362 0.451 1.000 1.000 BFAC 55.482 ATREF AX ALL AY ALL AZ ALL OCC 1 3 5 7 9 11 13 15 17 19 AOCC 1 3 5 7 9 11 13 15 17 19 BFAC 2 4 6 8 10 12 14 16 18 20 END-phare cad |
CCP4 document for
truncate
The native and derivative data sets are processed with truncate. Truncate converts the intensities into structure factors by taking the square root. Hence, the input files have the extension _i.mtz and the output files have the extension _f.mtz. It also puts the data sets on an absolute scale. This makes it convenient for estimating heavy atom occupancies. LABOUT -Output labels for each column of data. These should be meaningful and easy to remember. NRESIDUES -The number of residues in the asymmetric unit. Used to put the data sets on an absolute scale. ---------Logfile from truncate----------
Truncate outputs some useful statistics. The Wilson B-factor is calculated and plotted: SCALE = exp( - intercept). Least squares straight line gives: B = 21.434 SCALE = 216.53 where F(absolute)**2 = SCALE*F(observed)**2*EXP(-B*2*SINTH**2/L**2) |
|
|
|
|
|
| #!/bin/csh
-f
# dm hklin mtz/derivs_ph.mtz hklout mtz/dm.mtz << my-data >log/dm1.log SOLC 0.55 MODE SOLV HIST MULT COMBINE OMIT NCYCLE AUTO SCHEME ALL LABIN FP=Fnat SIGFP=sigFnat PHIO=PHIO FOMO=FOMO HLA=HLA HLB=HLB HLC=HLC HLD=HLD LABOUT PHIDM=PHI1 FOMDM=W1 HLADM=HLADM HLBDM=HLBDM HLCDM=HLCDM HLDDM=HLDDM my-data |
CCP4 document for
truncate
The native and derivative data sets are processed with truncate. Truncate converts the intensities into structure factors by taking the square root. Hence, the input files have the extension _i.mtz and the output files have the extension _f.mtz. It also puts the data sets on an absolute scale. This makes it convenient for estimating heavy atom occupancies. LABOUT -Output labels for each column of data. These should be meaningful and easy to remember. NRESIDUES -The number of residues in the asymmetric unit. Used to put the data sets on an absolute scale. ---------Logfile from truncate----------
Truncate outputs some useful statistics. The Wilson B-factor is calculated and plotted: SCALE = exp( - intercept). Least squares straight line gives: B = 21.434 SCALE = 216.53 where F(absolute)**2 = SCALE*F(observed)**2*EXP(-B*2*SINTH**2/L**2) |