UCLA Department of Chemistry and Biochemistry
153AH - Fall 2009 - Instructors: Todd Yeates, Duilio Cascio, Tobias Sayre
NmDsbA1: a protein from Neisseria meningitidis
by Howard Hsieh
|
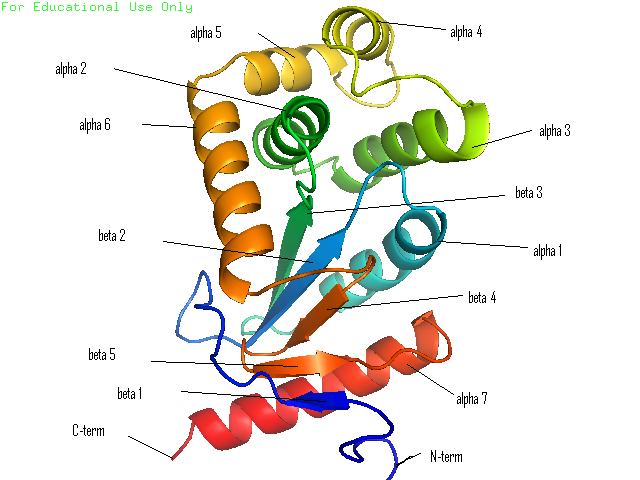
Many bacteria that are potentially pathogenic actually spend much of their time inhabiting human organs in a relatively benign state. Neisseria meningitidis is generally a commensal bacterium inside the nasopharynx of humans. However, it is capable of invading the mucosal epithelium and producing fatal meningitis and sepsis. In fact, N. meningitidis remains a significant worldwide health problem since it is responsible for thousands of deaths each year in Africa. N. meningitidis contains NmDsbA1, NmDsbA2 and NmDsbA3, which are three specific DsbA oxidoreductases that are vital for the oxidative folding of many membrane and secreted proteins. Therefore, removal of DsbA from N. Meningitidis is associated with loss of virulence. Among the three N. meningitidis DsbAs, NmDsbA1 and NmDsbA2 are lipoproteins anchored to the inner cell membrane; they share 78% amino acid sequence identity. NmDsbA3 is more divergent from NmDsbA1 and NmDsbA2 in that it only shares 57% and 51% amino acid sequence identity with the latter two proteins, respectively (1,2). The structure of NmDsbA1 is highly similar to known DsbA structures from other bacteria, especially E. coli. The DsbA fold is an extension of the thioredoxin superfamily and has two domains: a thioredoxin domain and a helical domain. The thioredoxin domain comprises a β-sheet (strands β2, β3, β4 and β5) flanked by helices α1, α7 and the C-terminal portion of α6. The helical domain which is inserted into the thioredoxin domain consists of helices α2, α3, α4, α and the N-terminal portion of α6 (Figure 1). NmDsbA1 crystallized with six molecules in the asymmetric unit and all of them are nearly identical with root-mean-square distance (rmsd) values over all Cα positions for each chain compared to chain A ranging from 0.39 Å to 0.54 Å (1). The molecules are arranged as head to tail repeats related by 180°. The head to tail arrangement allows the N-terminus of one protein to bind the peptide-binding cleft of a neighboring molecule. The N-terminal peptide region comprises vector-derived residues -4 to -1 (Asp-Asp-Asp-Lys). This charged sequence makes a series of polar and non-polar contacts within the binding cleft (Figure 2). The reactive disulfide from NmDsbA1 can be found in each of the six crystal copies. However, the active-site cysteines are modeled in different conformations because the two conformations of the active-site cysteines are consistent with the presence of both reduced NmDsbA1 and oxidized NmDsbA1 in the crystal (Figure 3). Even though the function of NmDsbA1 has not been elucidated fully, it is believed that NmDsbA1 depends on a pair of reactive cysteines separated by two residues (the CPHC motif) in order to invade the host. Therefore, future work could focus on the disulfide bonds between cysteines for the purpose of deactivating NmDsbA1. References (1) Vivian, J.P.,et al. (2009). Structure and function of the oxidoreductase DsbA1 from Neisseria meningtdis, J. Mol. Biol. doi:10.1016/j.jmb.2009.09.065 (2) Sinha, S.et al. (2008). Reduced DNA binding and uptake in the absence of DsbA1 and DsbA2 of Neisseria meningitidis due to inefficient folding of the outer-membrane secretin PilQ. Microbiology 154, 217-225 (3) PDB code: 3A3T |
|