UCLA Department of Chemistry and Biochemistry
153AH - Fall 2009 - Instructors: Todd Yeates, Duilio Cascio, Tobias Sayre
Phosphoenolpyruvate Carboxylase: A Homotetrameric Enzyme in Maize
by Priscilla Sugianto
|
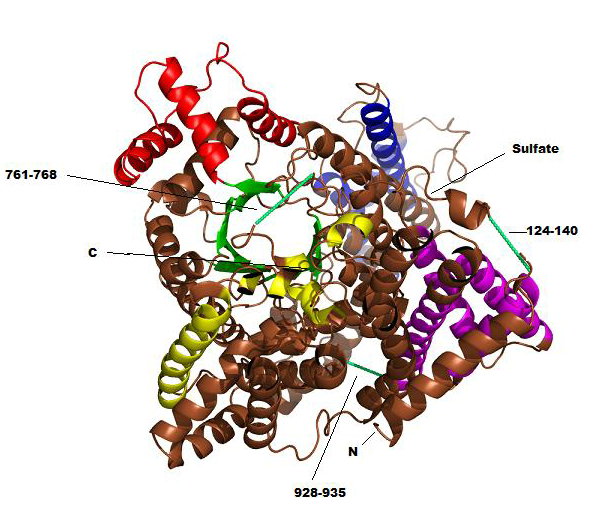
Phosphoenolpyruvate carboxylase (PEPC) is a homotetrameric regulatory enzyme that catalyzes the carboxylation of phosphoenolpyruvate (PEP) in the presence of bicarbonate (HCO3-) to form oxoloacetate (OAA) and phosphate (PO43-). It is found in all photosynthetic organisms, including C4 plants and a variety of photosynthetic bacteria, and is crucial in the fixation of atmospheric CO2 during photosynthesis. All known PEPCs show significant conservation, suggesting that reaction mechanisms among the enzymes from various organisms are essentially the same. PEPC is allosterically activated by glucose-6-phosphate (G6P) in all plants and by glycine in monocot plants (e.g., maize), and it is negatively controlled by malate and aspartate. Despite the knowledge of the crystal structure, the overall mechanism for the carboxylation and allosteric regulation remains unclear. The overall structure of tetrameric maize PEPC (ZmPEPC) appears to be a dimer of dimers (Fig. 1). Each monomer contains an 8-stranded β-barrel along with approximately 40 α-helices (Fig. 2). The tetramer has an overall size of approximately 130 x 120 x 70 Å. The two monomer chains of ZmPEPC is the asymmetric unit of the crystal are related by a non-crystallographic 2-fold axis. The secondary structural features show that helices comprise 65% of the polypeptide, whereas β strands comprise only 5%. The N-terminal region (residues 1-34) and three loops comprising residues 124-140, 761-768, and 928-935 show little or no electron density. Since there was no apparent proteolytic cleavage as judged by SDS-PAGE, it is likely that the loops exist in multiple conformations. Only the N-terminal region is partially truncated by proteolytic treatment. The removal of this N-terminal region makes the enzyme catalytically more active. The reported structure includes a sulfate ion at the putative binding site for an allosteric activator, G6P. In the vicinity of the ZmPEPC bundle of helices, a higher peak of electron density is surrounded by four positively charged residues (Arg183, Arg184, Arg231, and Arg372 in the neighbor subunit) (Figure 3). The extra density is large enough to accommodate a sulfate, which is located at the N terminus of the α7 helix in bundle A. In addition, the side chain of Arg372 interacts with the side chain of Ser185, which appears to be significant for the allosteric regulation mechanism. Since a pocket is present around the site where sulfate was observed, this site may be the binding site for G6P. Further studies from three-dimensional structure, combined with related biochemical studies, may clarify the mechanisms for the carboxylation reaction and for the allosteric regulation of PEPC. References Matsumura, H, et al. (2002). Crystal Structures of C4 Form of Maize and Quaternary Complex of E. coli Phosphoenolpyruvate Carboxylases. Structure 10, 1721-30. Kai, Y, Matsumura, H., and Izui, K. (2003). Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 414, 170-9. PDBID 1JQO |
|