UCLA Department of Chemistry and Biochemistry
153AH - Fall 2009 - Instructors: Todd Yeates, Duilio Cascio, Tobias Sayre
Hemagglutinin of influenza A virus in complex with human antibody CR6261 Fab
by Phoebe Lao
|
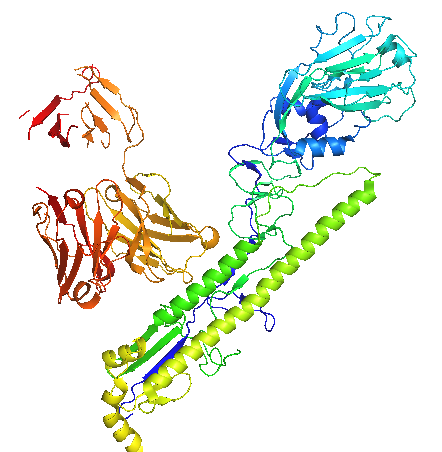
Human influenza A pandemics have swept the world in the past century. Each pandemic virus is derived, to some degree, from an avian influenza virus. Because of the sheer number and different combination of influenza A subtypes, predicting a subtype and thus developing vaccine ahead of time is virtually impossible. Therefore, vaccines that stimulate the production of antibodies that neutralize different subtypes would be immensely useful. The flu virus Hemagglutinin, or HA is the major contributor to the fusion of virus and human cell membranes, allowing the flu virus to insert its genetic material into the human cell. HA is cleaved into HA1 and HA2 during viral maturation. It has been determined that the HA2 is highly conserved across many different subtypes, and that human antibody CR6261 neutralizes these subtypes by interacting with HA2, which is comprised primarily from two helices: A and CD (Fig. 1)(1). The structure of the CR6261 Fab antibody fragment in complex with HA of H1N1/H5N1 has been elucidated in order to show the mechanism of neutralization (1). HA1 is the membrane-distal part, with a globular head and C- and N-termini extended toward HA2. HA2 is the major site of interaction with the CR 6261 epitope, which consists of a heavy chain on top of a light chain (1). A universally-conserved Asn154 (Fig.2) on helix A prevents its interaction with the light chain, so CR6261 interacts mainly with the heavy chain, which makes hydrogen bonds with helix A through its heavy chain complementary determining regions (HCDR's) (Fig.3). Five of the hydrogen bonds are conserved between H1N1 and H5N1 viruses, suggesting the importance of the conserved helix A as the major component of interaction across different subtypes. In contract, HCDR's only make hydrophobic contacts with HA1 (1). The structure of the complex also reveals the mechanism of binding to host cell membranes and how CR6261 prevents it. A pH-dependent rearrangement is thought to be the major reason for the conformational change in HA that initiates the membrane fusion process (1). At low pH, the association between HA1 and HA2 weakens, allowing HA1 to pivot and adopt a different conformation, which destabilizes HA2. HA2 is also sensitive to low pH; the loop between helix A and CD forms an α-helix that pulls the viral and target membranes close together. Being positioned just below the globular head of HA1, CR6261 prevents the pivoting and therefore stabilizes HA in the pre-fusion conformation, effectively preventing fusion of the viral and host cell membranes (1). CR6261 neutralizes many subtypes, though not subtypes H3 and H7. This can be explained by the amino acids constitution of helix A. The CR6261 sites on HA2 are almost invariant from one subtype to another. However, His38 in the H1 epitope, sitting directly across from the Fab heavy chain, is substituted by Asn38 in the H3 subtype. Asn38 is subjected to N-linked glycosylation and therefore abolishes CR6261 binding to HA2. In fact, introducing Asn38 into H5N1 reduces the CR6261 binding by approximately 70% (1). Despite its limited ability to neutralize certain subtypes of influenza A, CR6261 can recognize the conserved HA of many other subtypes. The complex of CR6261 and HA of H1N1 and H5N1 provides valuable information on the antibody’s mechanism of neutralization. It also offers new leads toward developing a heterosubtypic vaccine against influenza A. References (1) Ekiert, et al. (2009). Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246-251. (2) PDBID 3GBN |
|