UCLA Department of Chemistry and Biochemistry
153AH - Fall 2009 - Instructors: Todd Yeates, Duilio Cascio, Tobias Sayre
Prions: Infectious Proteins with Mysterious Propagation
by Hannah Mendoza
|
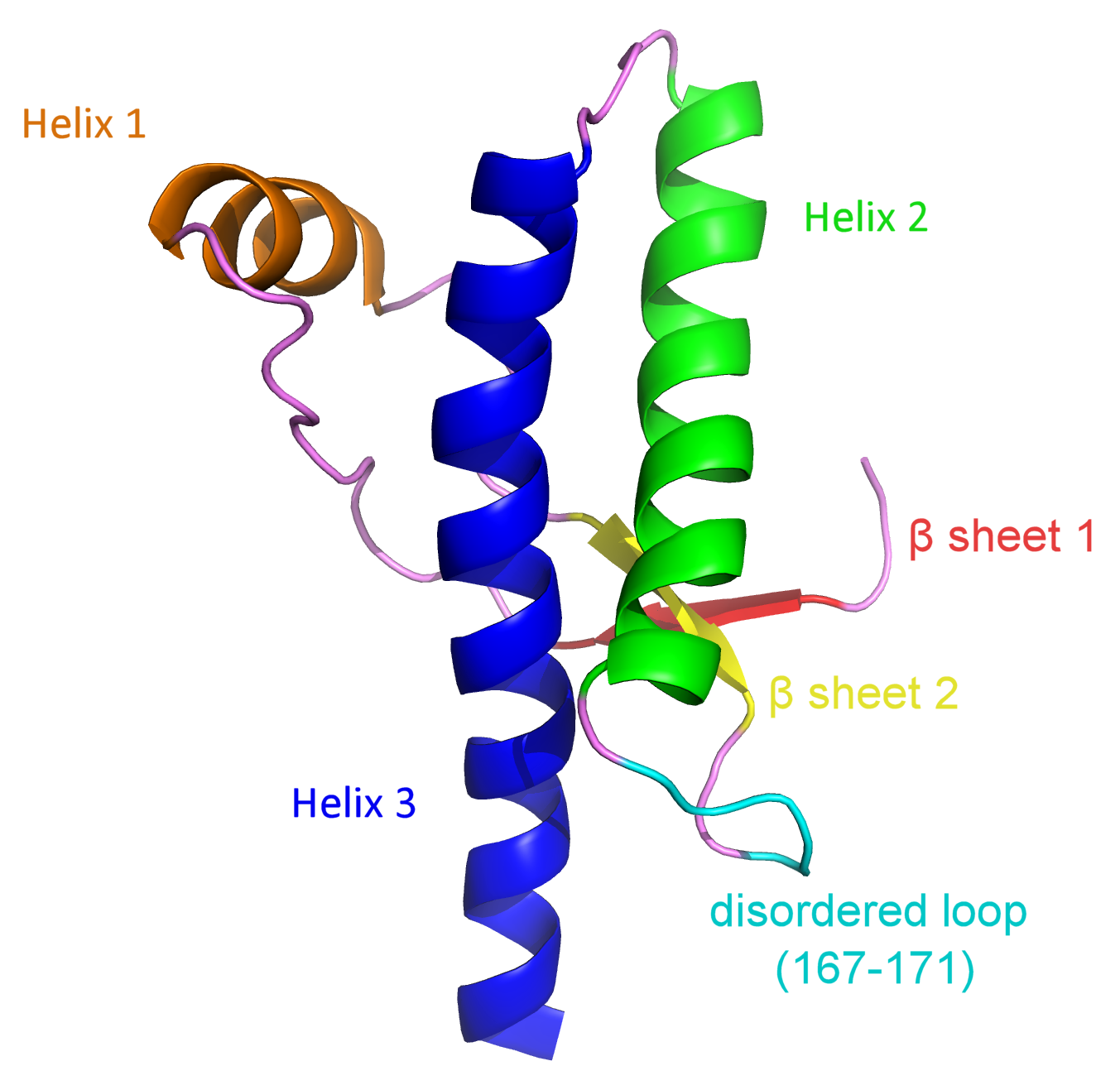
Prions are infectious particles believed to be composed entirely of protein. They propagate disease by inducing normal prion proteins found in the brain to misfold, causing severe brain damage and disease. Such diseases include Bovine Spongiform Encephalopathy (BSE or mad cow disease), Scrapie, and Creutzfeldt Jakob disease (CJD). Researchers have not yet proven the mechanism by which prions operate. Hypotheses have been put forward to explain the ability of prions to induce the pathological conformation change, including the "protein-only hypothesis." It theorizes that the proteins in the abnormal prion state have the ability to convert normal prions into the abnormal conformation without the need for a nucleic acid sequence. This was a ground breaking idea because it challenged the central dogma that nucleic acids are the only biological molecules that can store and replicate genetic information. Normal cellular prion proteins, noted as PrPC, function in the brain on the surface of nerve cells. Although their exact purpose is not known, prions have been reported to maintain long-term memory, cell to cell adhesion, and cell communication. Abnormal prions, denoted as PrPSc, transform PrPC in a complex manner still not fully understood. However, it is known that PrPSc is highly infectious; in principle, only a few copies of the molecule could convert a substantial fraction of the PrPC in an organism. PrPC is a monomeric protein that is mostly α-helical and protease sensitive. The structure of human PrPC contains residues 23-230 with a highly disordered N-terminus (23-124) and a globular C-terminal domain consisting of three α-helices (144-154, 173-194, and 200-228) and two β-sheets (128-131 and 161-164). The sites most interesting on the protein are the highly disordered loop at residues 167-171 (colored cyan in fig. 1) and the region of helix 3 (colored blue in fig. 1) in acidic conditions. Moreover, structural studies have discovered that at physiological pH (around 6.5-7.8), octapeptide repeats (OPRs) located in the N-terminus are highly ordered. These OPRs, found in residues 61-90, encompass the amino acid sequence PHGGGWGQ and are highly conserved among mammalian prion proteins. Structural data suggest that pH-dependent aggregation is due to homo-oligomeric interactions between the four OPRs. This type of aggregation occurs naturally in neuron cells and promotes the concentration of PrPC at the presynaptic membrane surface of neurons, stimulating PrPC endocytosis into vesicles. Additionally, the OPRs possibly serve as a site for cell adhesion between axons and dendrites, contributing to neuronal cell communication. While structural studies using NMR gave additional insight into the structure of PrPC, it has not been possible to obtain the complete structure of PrPSc. PrPSc is known to have a higher concentration of beta sheets compared to PrPC (45% compared to 3%), thereby contributing to its fibrous conformation. Fig. 2 shows fibrils of a fungus HET-s prion domain, whose β-solenoid fold has been proposed for the structure of PrPSc. Characterizing PrPSc structurally has been challenging, due to the difficulty in studying it through NMR or X-ray crystallography methods. References Wasmer, et al. (2008). Amyloid fibrils of the HET-s(218–289) prion form a β-solenoid with a triangular hydrophobic core. Science 319, 1523-1526. Harris, et al. (2006). New insights into prion structure and toxicity. Neuron 50, 353-357. Zahn, et al. (2003). The Octapeptide repeats in mammalian prion protein constitute a pH-dependent folding and aggregation site. J. Mol. Biol. 334(3):477-88. PDBID: 1qm2 PDBID: 2rnm |
|