UCLA Department of Chemistry and Biochemistry
153AH - Fall 2009 - Instructors: Todd Yeates, Duilio Cascio, Tobias Sayre
IRF-1: A Regulator of Interferons in Innate Immune Response
by Allison Truong
|
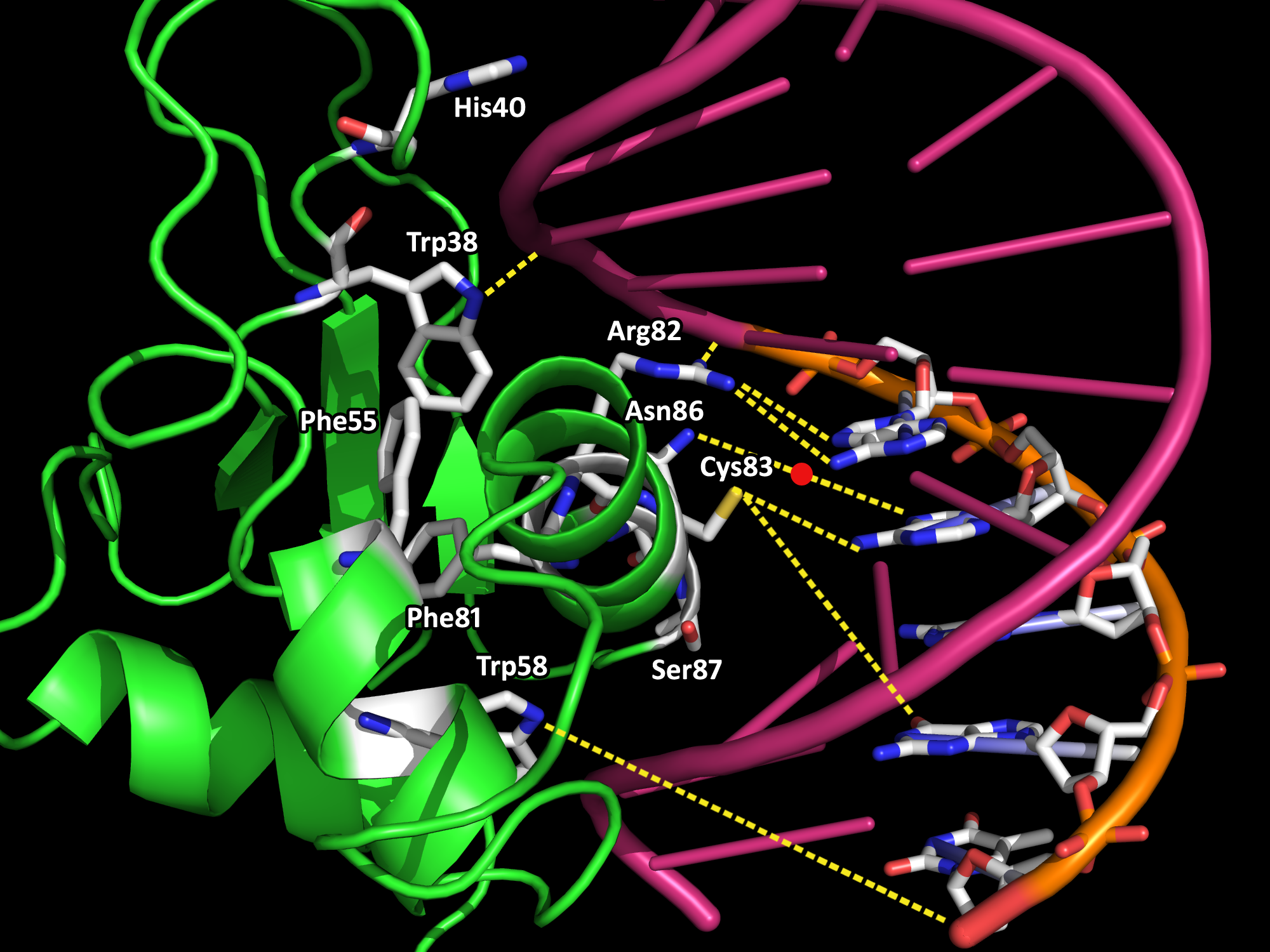
The innate immune system is the body's first line of defense against microbial pathogens. The induction of gene transcription in response to extracellular stimuli constitutes an important component of host defense mechanisms. IRF-1 is a mammalian transcription factor protein belonging to a family of interferon regulatory factors (IRF) that regulates interferon production, interferon-inducible genes, cell growth and apoptosis, and tumor suppression (3,4). Interferons (IFN) are families of glycoproteins known as cytokines that provide the host with protective defenses against pathogens including viruses, bacteria, and parasites, and against tumor cells (4). IRF-1 is constitutively localized in the nucleus. It is roughly 30 Å in diameter and 113-residues long (Fig. 1). IRF-1 possesses a helix-turn-helix motif with a cluster of three α-helices and four β-sheets. The α+β architecture of the of the protein is interrupted by multiple loops that connect the various α/β or α/α secondary structures. Both the α-helices and the loops form extensive interactions with the DNA backbone via hydrogen bonds and van der Waal forces. Upon DNA binding, the IRF-1 loops become ordered, which results in a decrease in entropy that is thermodynamically satisfied via the loss of water molecules and cations on the DNA backbone (1). Water release from protein-DNA interfaces upon binding contributes favorably to the energetics of binding, while the replacement of cations from the negatively charged surface of the DNA with positively charged side chain groups from the interacting protein is also entropically favored (2). Crystal structures have been determined of many proteins within the IRF family. They are characterized by a distinctive 'tryptophan cluster' DNA-binding region (Fig. 2). The IRF-1 sequence is specific and unique compared to other transcription factors except for the conservation of three of the five characteristic tryptophan residues that contact the DNA bases in the major groove. There is an important coupling between the hydrophobic interior core of IRF-1 due to the interactions between the tryptophan and phenylalanine residues and the DNA backbone (Fig. 3). A single mutation in any of the tryptophans may lead to the instability of IRF-1 and the inability of IRF-1 to bind to DNA (1). The IRF-1 protein was isolated on the basis of its affinity to DNA elements of the natural positive regulatory domain I and III (PRD I/III) from the interferon-β promoter that mediates virus responsiveness (1). PRD I/III of the interferon-β enhancesome possesses a short GAAA DNA-base-pair core sequence that bends approximately 22° toward IRF-1 upon the formation of the IRF-1-DNA complex (1). The shape of IRF-1 is tangential to the DNA sugar-phosphate backbone, providing an optimal angle of attack for making extensive interactions between the protein and DNA. Future research directions include mutagenesis studies aimed at studying IRF-1 and DNA interactions, as well as functional studies of IRF-1 and other transcriptional activators on the promoters of distinct genes under different physiological conditions. References (1) Escalante, et al. (1998). Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature 391, 103-106. (2) Jayaram, B., Jain, T. (2004). The Role of Water in Protein-DNA Recognition. Annual Rev. Biophys. Biomol. Struct. 33, 343-361. (3) Kröger, et al. (2002). Activities of IRF-1. Journal of Interferon & Cytokine Research 22, 5-14. (4) Taniguchi, et al. (1995). Regulation of the interferon system and cell growth by the IRF transcription factors. J. Cancer Res. Clin. Oncol. 121, 516-520. (5) PDBID 1if1 |
|