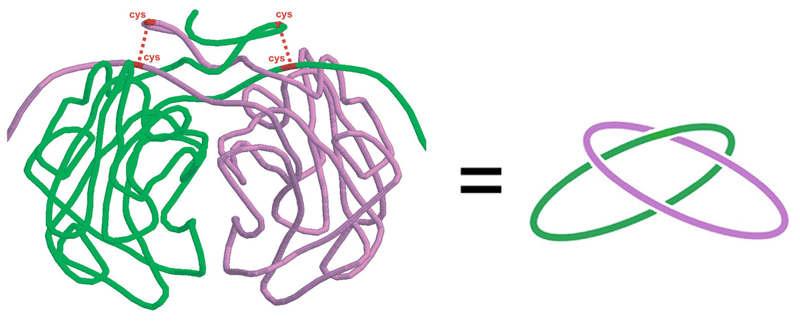
| Although proteins often adopt rather complex folds, the fact of the matter is that the backbone of a protein molecule adopts a knotted configuration in only a very few proteins among the thousands of known protein folds. This is contrary to what would be expected if protein backbones followed essentially random paths in space; for spatial curves as long as medium-sized proteins, knots would be expected to dominate. The observation that native protein folds hardly sample the space of knotted conformations - a space that represents the majority of all possible conformations - has profound implications for protein folding in general, and for the Thermodynamic Hypothesis (first posed by Anfinsen) in particular. There are however a few special cases where knotted proteins have been observed, and these provide special opportunities for studying protein folding and stability. The intuition that it might be kinetically difficult for a knotted protein chain to reach its native state can be considered in reverse, with the conclusion that knotting and other kinds of topological entaglements could kinetically stabilize the folded states of proteins containing these features. While looking for unusual topological features in protein stuctures, Neil King recently discovered a novel slipknot feature in a well-known thermostable protein, alkaline phosphatase. Beyond the knotting of individual protein chains, a few rare cases are known in which protein chains from two different subunits form topologically interlinked chains. One of these was discovered by Danny Boutz in the citrate synthase dimer from P. aerophilum. Experimental work and design studies on topologically complex proteins are in early stages of exploration. |  |
Boutz, D.R., Cascio, D., Whitelegge, J., Perry, L.J., and Yeates,
T.O. (2007).
Discovery of a thermophilic protein complex
stabilized by topologically interlinked chains.
J. Mol. Biol. 368,
1332-44.
Norcross, T.S. and Yeates, T.O.
2006. A framework for
describing topological frustration in models of protein folding. J. Mol. Biol. 362, 605-21.
King, N.P., Yeates,
E.O., Yeates, T.O. (2007). Identification
of Rare Slipknots in Proteins and Their Implications for Stability and Folding.
J. Mol. Biol. 373, 153-66.