|
Racemic protein crystallography refers to the idea of crystallizing
proteins from a racemic mixture of the natural, biologically-handed,
molecule and its mirror image molecule (reviewed in Yeates and Kent, 2012). The latter
must be chemically synthesized in the laboratory from D-amino
acids while the natural molecule may be synthezised or expressed in a biological host by
using traiditional molecular biology methods. Laura Zawadzke
and Jeremy Berg were the first to execute the idea in 1993 using
the small (45 amino acid) protein rubredoxin. An early motivation
for pursuing such studies was the idea that structure determination might be easier or more robust
using diffraction data from a centrosymmetric crystal, which requires growth
from a racemic mixture. There are merits to this idea, and recent
work has further explored the possibilities of certain advantages in phasing
centrosymmetric protein diffraction data (Sawaya et al., 2012).
Setting aside possible advantages in phasing and structure determination that might arise
with centrosymmetric protein crystals, there is now good reason to believe that racemic
crystallography could have a much more profound impact, by dramatically improving
the ease with which macromolecular crystals can be obtained; the crystallization problem remains the most
serious and most vexing obstacle in macromolecular crystallography. The prediction in 1995 that
crystallization from a racemic mixture might provide a critical advantage arose largely as a surprise
while working on
a mathematical explanation for why some special crystal space groups are preferred so strongly
over others by proteins when they crystallize (i.e. in ordinary experiments involving only the natural biological hand).
The preferences for different crystal space groups
vary by more than two orders of magnitude, so understanding that phenomenon has potentially
important implications for the crystallization problem as a whole. The paper
by Stephanie Wukovitz in 1995 offered
a mathematical explanation for the space group preference phenomenon; i.e. it explained why
P212121 is by far the most commonly observed crystal space group for (ordinary chiral) proteins.
Reaching farther, it was also noted at that time that the theory predicted that even more
dominant crystal space groups existed among those that are only possible when using a
racemic mixture. Two predictions were made: that proteins would crystallize with greater ease
from racemic mixtures owing to the existence of especially favored racemic crystal symmetries,
and that P1(bar) would be the dominant space group observed.
The invention of native chemical ligation methods by Phil Dawson and Stephen Kent in the early 1990's
opened up prospects for chemically synthesizing larger protein molecules. Kent and co-workers have
since tested racemic crystallography on a wide range of protein molecules. Current data provide strong support
for the idea that proteins do crystalize with realtive ease from synthetic racemic mixtures (and most often
in P1(bar) as predicted).
References:
- Yeates TO, Kent SB. (2012).
Racemic protein crystallography.
Annu Rev Biophys. 2012. 41:41-61.
[Abstract]
Although natural proteins are chiral and are all of one "handedness," their mirror image forms can be prepared by chemical synthesis. This opens up new opportunities for protein crystallography. A racemic mixture of the enantiomeric forms of a protein molecule can crystallize in ways that natural proteins cannot. Recent experimental data support a theoretical prediction that this should make racemic protein mixtures highly amenable to crystallization. Crystals obtained from racemic mixtures also offer advantages in structure determination strategies. The relevance of these potential advantages is heightened by advances in synthetic methods, which are extending the size limit for proteins that can be prepared by chemical synthesis. Recent ideas and results in the area of racemic protein crystallography are reviewed.
- Sawaya MR, Pentelute BL, Kent SB, Yeates TO. (2012).
Single-wavelength phasing strategy for quasi-racemic protein crystal diffraction data.
Acta Crystallogr. D Biol. Crystallogr.. Jan 2012. 68(Pt 1):62-8.
[Abstract]
Racemic protein crystallography offers two key features: an increased probability of crystallization and the potential advantage of phasing centric diffraction data. In this study, a phasing strategy is developed for the scenario in which a crystal is grown from a mixture in which anomalous scattering atoms have been incorporated into only one enantiomeric form of the protein molecule in an otherwise racemic mixture. The structure of a protein crystallized in such a quasi-racemic form has been determined in previous work [Pentelute et al. (2008), J. Am. Chem. Soc. 130, 9695-9701] using the multiwavelength anomalous dispersion (MAD) method. Here, it is shown that although the phases from such a crystal are not strictly centric, their approximate centricity provides a powerful way to break the phase ambiguity that ordinarily arises when using the single-wavelength anomalous dispersion (SAD) method. It is shown that good phases and electron-density maps can be obtained from a quasi-racemic protein crystal based on single-wavelength data. A prerequisite problem of how to establish the origin of the anomalous scattering substructure relative to the center of pseudo-inversion is also addressed.
- Banigan JR, Mandal K, Sawaya MR, Thammavongsa V, Hendrickx AP, Schneewind O, Yeates TO, Kent SB. (2010).
Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci.. Oct 2010. 19(10):1840-9.
[Abstract]
The 50-residue snake venom protein L-omwaprin and its enantiomer D-omwaprin were prepared by total chemical synthesis. Radial diffusion assays were performed against Bacillus megaterium and Bacillus anthracis; both L- and D-omwaprin showed antibacterial activity against B. megaterium. The native protein enantiomer, made of L-amino acids, failed to crystallize readily. However, when a racemic mixture containing equal amounts of L- and D-omwaprin was used, diffraction quality crystals were obtained. The racemic protein sample crystallized in the centrosymmetric space group P2(1)/c and its structure was determined at atomic resolution (1.33 A) by a combination of Patterson and direct methods based on the strong scattering from the sulfur atoms in the eight cysteine residues per protein. Racemic crystallography once again proved to be a valuable method for obtaining crystals of recalcitrant proteins and for determining high-resolution X-ray structures by direct methods.
- Pentelute BL, Mandal K, Gates ZP, Sawaya MR, Yeates TO, Kent SB. (2010).
Total chemical synthesis and X-ray structure of kaliotoxin by racemic protein crystallography.
Chem. Commun. (Camb.). Nov 2010. 46(43):8174-6.
[Abstract]
Here we report the total synthesis of kaliotoxin by 'one pot' native chemical ligation of three synthetic peptides. A racemic mixture of D- and L-kaliotoxin synthetic protein molecules gave crystals in the centrosymmetric space group P1 that diffracted to atomic-resolution (0.95 Å), enabling the X-ray structure of kaliotoxin to be determined by direct methods.
- Wukovitz SW, Yeates TO. (1995).
Why protein crystals favour some space-groups over others.
Nat. Struct. Biol.. Dec 1995. 2(12):1062-7.
[Abstract]
One of the most puzzling observations in protein crystallography is that the various space-group symmetries occur with striking non-uniformity. Molecular close-packing has been invoked to explain similar observations for crystals of small organic compounds, but does not appear to be the dominant factor for proteins. Instead, we find that the observed frequencies for both two- and three-dimensional crystals can be explained by an entropic model. Under a requirement for connectivity, the favoured space groups are simply less restrictive than others in that they allow the molecules more rigid-body degrees of freedom and can therefore be realized in a greater number of ways. This result underscores the importance of the nucleation event in crystallization and leads to specific ideas for crystallizing water-soluble and membrane proteins.
|
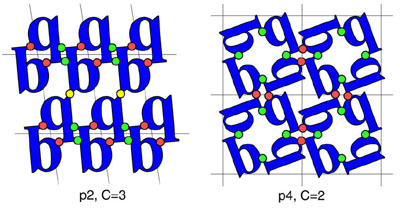
A diagram illustrating the minimum contact number, C, for two layer groups.
Determining these values for the 65 3D space groups made it possible
to define the number of rigid body degrees of freedom, D, available to a collection of molecules
subjected to being arranged in a particular space group. This quantity explains much of the dramatic variation in
observed space group preferences for proteins. An extension of the idea to the racemic space groups,
which are generally inaccessible to chiral biological molecules, suggested that proteins could be crystallized with
much greater ease from a (synthetic) racemic mixture.
(Adapted from Wukovitz and Yeates, 1995).
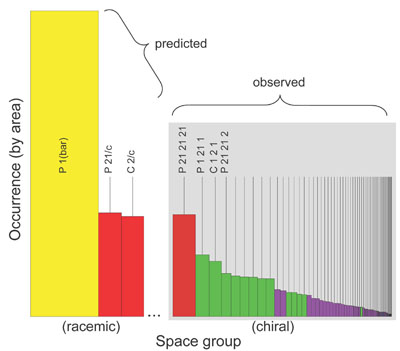
A histogram showing the observed occurrence of space groups for protein
crystals (right). Each bar represents the occurrence of a single space group.
The panel on the right shows
the 65 'biological' space groups (presented in their standard numerical order) colored according to their 'dimensionality', according to Wukovitz and Yeates
(red: D=7 [only P212121]; green: D=6; purple: D=5; blue: D=4 [too rare to be easily visible]).
The value of D, computed from purely mathematical quantities related to space group symmetry, divides the 65 space groups
into nearly non-overalapping preference categories. The panel on the left shows a rough prediction of what might be
expected for crystallization from racemic protein samples. P1(bar) -- the only space group out of 230 for which D=8 --
has been predicted to dominate (see Wukovitz and Yeates, 1995), while P21/c and C2/c are also expected to be common.
(Adapted from Yeates and Kent, 2012).
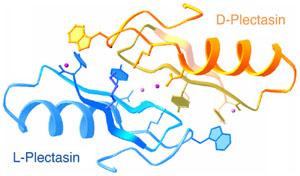
(left) An example of a protein structure in a racemic crystal form in space group P1(bar).
(Adapted from Yeates and Kent, 2012).
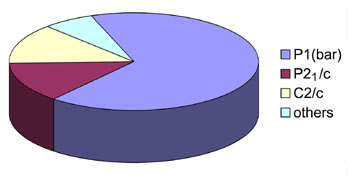
(right) A pie chart showing the observed racemic space groups for protein crystals based on about 15 crystals obtained
up until 2012. The emerging trend clearly confirms the prediction from theory in 1995.
(Adapted from Yeates and Kent, 2012).
|