|
The toughest obstacle in determining the three-dimensional structures
of macromolecules is obtaining crystals of the molecule in
question -- protein or nucleic acid.
The process of crystallization remains largely an art form. In favorable cases,
success usually requires
very many experimental trials under different conditions. In unfavorable
cases, crystals are never obtained. The reasons for failure are sometimes
understandable -- e.g. flexibility or heterogeneity -- but are just as often
unknown. Numerous strategies have been developed to improve the chances of
success. Some of these are based on the principle that it is more
fruitful to create variations on the macromolecule of interest, and to attempt
crystallizing these multiple variants, rather
than performing an infinitude of different trials on a single molecular
construct.
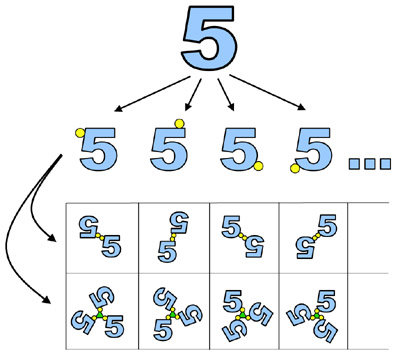
One family of approaches, which we refer to as 'synthetic symmetrization' (Banatao, et al., 2006),
seeks to make a series of distinct symmetric constructs from a starting
macromolecule that is otherwise asymmetric (i.e. a monomer)
(see Figure at right). If the starting molecule can be made
symmetric in a variety of distinct ways (e.g. by dimerization
at different contact points), then each
of the resulting constructs will enjoy distinct opportunities for packing in
crystalline arrangements. In addition, the presence of symmetry, by itself, confers some
advantage in crystallization.
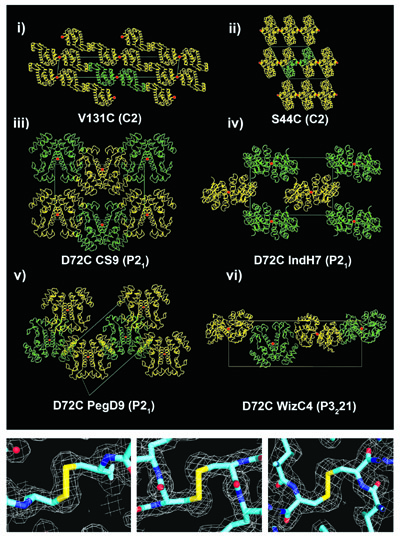
The first approach demonstrated for synthetic symmetrization was disulfide
bonding following the introduction of single cysteine residues at surface positions in
a protein molecule. Applying the
method to lysozyme led to six new crystal forms of that protein (Banatao, et al., 2006).
The cysteine cross-linking
approach was subsequently used to determine the structure of a protein of previously unknown
structure, endoglucanase A (Forse, et al., 2011).
A potentially more versatile strategy based on metal binding was developed by three graduate students
(Laganowsky, Zhao, Soriaga, et al., 2011), adapting ideas introduced by Akif Tezcan's laboratory.
Two histidine residues are introduced at positions i and i+4 in a region of the sequence
predicted to constitute a surface-exposed helix.
During crystallization, addition of metal (i.e. nickel, zinc, or copper) leads to
formation of dimers (or sometimes other oligomeric species). Distinct constructs,
with histidines introduced at different positions, have distinct crystal packing opportunities
and give rise to new crystal forms. The metal-based symmetrization approach has so far
been demonstrated with lysozyme and maltose binding
protein as model systems.
References:
- Laganowsky A, Zhao M, Soriaga AB, Sawaya MR, Cascio D, Yeates TO. (2011).
An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci.. Nov 2011. 20(11):1876-90.
[Abstract]
Combining the concepts of synthetic symmetrization with the approach of engineering metal-binding sites, we have developed a new crystallization methodology termed metal-mediated synthetic symmetrization. In this method, pairs of histidine or cysteine mutations are introduced on the surface of target proteins, generating crystal lattice contacts or oligomeric assemblies upon coordination with metal. Metal-mediated synthetic symmetrization greatly expands the packing and oligomeric assembly possibilities of target proteins, thereby increasing the chances of growing diffraction-quality crystals. To demonstrate this method, we designed various T4 lysozyme (T4L) and maltose-binding protein (MBP) mutants and cocrystallized them with one of three metal ions: copper (Cu²⁺, nickel (Ni²⁺), or zinc (Zn²⁺). The approach resulted in 16 new crystal structures--eight for T4L and eight for MBP--displaying a variety of oligomeric assemblies and packing modes, representing in total 13 new and distinct crystal forms for these proteins. We discuss the potential utility of the method for crystallizing target proteins of unknown structure by engineering in pairs of histidine or cysteine residues. As an alternate strategy, we propose that the varied crystallization-prone forms of T4L or MBP engineered in this work could be used as crystallization chaperones, by fusing them genetically to target proteins of interest.
- Forse GJ, Ram N, Banatao DR, Cascio D, Sawaya MR, Klock HE, Lesley SA, Yeates TO. (2011).
Synthetic symmetrization in the crystallization and structure determination of CelA from Thermotoga maritima.
Protein Sci.. Jan 2011. 20(1):168-78.
[Abstract]
Protein crystallization continues to be a major bottleneck in X-ray crystallography. Previous studies suggest that symmetric proteins, such as homodimers, might crystallize more readily than monomeric proteins or asymmetric complexes. Proteins that are naturally monomeric can be made homodimeric artificially. Our approach is to create homodimeric proteins by introducing single cysteines into the protein of interest, which are then oxidized to form a disulfide bond between the two monomers. By introducing the single cysteine at different sequence positions, one can produce a variety of synthetically dimerized versions of a protein, with each construct expected to exhibit its own crystallization behavior. In earlier work, we demonstrated the potential utility of the approach using T4 lysozyme as a model system. Here we report the successful application of the method to Thermotoga maritima CelA, a thermophilic endoglucanase enzyme with low sequence identity to proteins with structures previously reported in the Protein Data Bank. This protein had resisted crystallization in its natural monomeric form, despite a broad survey of crystallization conditions. The synthetic dimerization of the CelA mutant D188C yielded well-diffracting crystals with molecules in a packing arrangement that would not have occurred with native, monomeric CelA. A 2.4 Å crystal structure was determined by single anomalous dispersion using a seleno-methionine derivatized protein. The results support the notion that synthetic symmetrization can be a useful approach for enlarging the search space for crystallizing monomeric proteins or asymmetric complexes.
- Banatao DR, Cascio D, Crowley CS, Fleissner MR, Tienson HL, Yeates TO. (2006).
An approach to crystallizing proteins by synthetic symmetrization.
Proc. Natl. Acad. Sci. U.S.A.. Oct 2006. 103(44):16230-5.
[Abstract]
Previous studies of symmetry preferences in protein crystals suggest that symmetric proteins, such as homodimers, might crystallize more readily on average than asymmetric, monomeric proteins. Proteins that are naturally monomeric can be made homodimeric artificially by forming disulfide bonds between individual cysteine residues introduced by mutagenesis. Furthermore, by creating a variety of single-cysteine mutants, a series of distinct synthetic dimers can be generated for a given protein of interest, with each expected to gain advantage from its added symmetry and to exhibit a crystallization behavior distinct from the other constructs. This strategy was tested on phage T4 lysozyme, a protein whose crystallization as a monomer has been studied exhaustively. Experiments on three single-cysteine mutants, each prepared in dimeric form, yielded numerous novel crystal forms that cannot be realized by monomeric lysozyme. Six new crystal forms have been characterized. The results suggest that synthetic symmetrization may be a useful approach for enlarging the search space for crystallizing proteins.
|
(left) A diagram illustrating the idea of 'synthetic symmetrization'
(Adapted from Banatao, et al., 2006). Application of disulfide-based symmetrization to T4 lysozyme led to
six new crystal forms (Adapted from Banatao, et al., 2006).
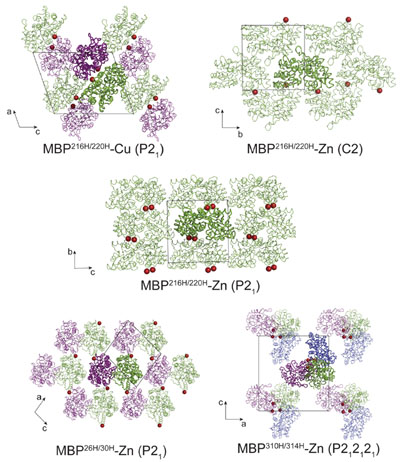
A diagram illustrating metal-based synthetic symmetrization. When applied to T4 lysozyme and MBP, the
method produced a total of 13 new crystal forms
(Adapted from Laganowsky, et al. 2011).
|