| Improved Twinning Server |
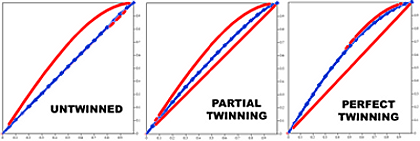
Is your crystal twinned? Merohedral twinning is an anomaly that is
impossible to detect by viewing the diffraction pattern. It occurs
when the lattices of two or more distinct crystal domains coincide
exactly in three dimensions. The twinning server can detect twinning
by comparing diffraction data to that expected by Wilson statistics.
If left undetected, you could spend months agonizing over poor phasing
results. You will save yourself untold mental anguish if you find out
now! The following crystallographic point groups support twinning: 3,
4, 6, 321, 312, 23.
Requirements: Unit cell parameters, space group, and observed structure factors in X-PLOR or CIF format. References: T. O. Yeates, Methods Enzymol. 276, 344-358, 1997. |
|
Matthews Coefficient
|
How many macromolecules are in your asymmetric unit? Are there
multiple molecules? A single molecule? Half a molecule? All these
are possible. The Matthews coefficient allows you to estimate this
number. The Matthews coefficient (Vm) is
easily calculated as
___________volume of your unit cell__________
Where Z is the number of asymmetric units in the unit cell (i.e. the number of symmetry operators in your space group). The unknown variable, X, is the number of molecules in the asymmetric unit. You may perform the calculation using the Vm server . Calculate a series of Matthews coefficients with X= 0.5, 1.0, 2.0, etc. The most probable values of X are those which give Matthews coefficients within the empirically observed range. This range is shown in the bar graph at left, or click on the reference below to see the original paper in PDF format. Requirements: Unit cell parameters, space group, molecular weight of your macromolecules. References: B. W. Matthews, J. Mol. Biol. 33, 491-497, 1968. |
| Self Rotation Function |
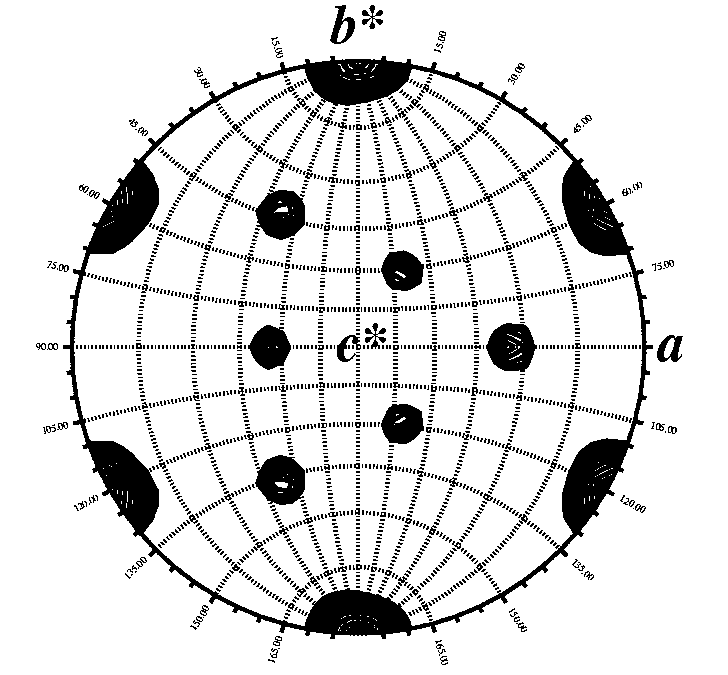
Did the Matthews Coefficient suggest 2 or more molecules in the
asymmetric unit? The self rotation function will tell you if the
molcules in your asymmetric unit are related by rotational symmetry
axis and if so, how the axis is oriented in your unit cell. This
information is crucial if you want to take advantage of
non-crystallographic symmetry (NCS) averaging.
Requirements: Unit cell parameters, space group, and observed structure factors in any format. References: M. G. Rossmann and D. M. Blow, Acta Cryst. 15, 24-31, 1962. |
| Native Patterson Map |
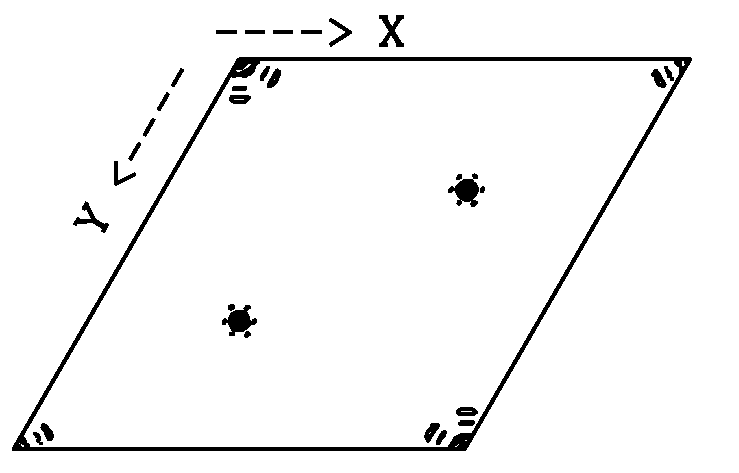
Did the Matthews Coefficient suggest 2 or more molecules in the
asymmetric unit? The native Patterson map will tell you if the
molcules in your asymmetric unit are related by a pure translation
and if so, the length and direction of the vector that relates the
two molecules. This
information is crucial if you want to take advantage of
non-crystallographic symmetry (NCS) averaging.
Requirements: Unit cell parameters, space group, and observed structure factors in XtalView's fin format.
References:
Reference for the Native Patterson Map:
|