|
Part One:
Crystallization Trials
|
Objective: To crystallize proteinase
K. You will optimize an initial set of crystallization conditions
discovered earlier by sparse matrix screening kits. This year, we are
optimizing the ratio of proteinase K volume to reservoir volume (3:1,2:1,
and 1:1).
Method: Hanging drop vapor diffusion.
Materials:
1) 3 mg of Proteinase K (ProK) from Tritirachium album purchased from Sigma
(cat. no. P2308).
2) 4M ammonium sulfate (NH4)2SO4
3) Tris buffer pH 8.0 1M
4) Distilled water
5) Greased VDX crystallization plates, 24-wells (Hampton Research
(cat. no. HR3-140).
6) Siliconized glass cover slips (Fisherbrand
Microscope Cover Glass 22x22-2 cat. no. 12-540B)
Procedures:
1) Work individually. Each person sets up a 24-well plate.
2) Dissolve 3 mg of ProK (supplied in 0.5 mL
microcentrifuge tube) in 100 uL of distilled
water. Using pipet tip, stir gently to dissolve the protein. No bubbles!
The final concentration of protein should be 30 mg/mL.
Aliquot 20 uL of ProK
into each of 4 empty microcentrifuge tubes. Draw the solution from the top
of the tube, not the bottom.
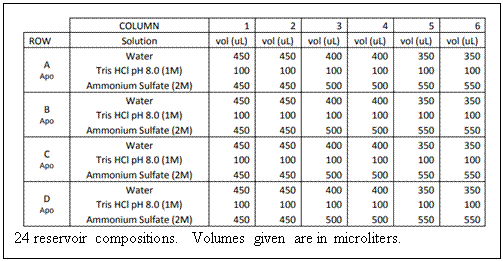 3) Prepare reservoir
solutions: Pipet the indicated amount of reagent to each of the 24
wells. [(NH4)2SO4] concentration
varies horizontally across a row. The volume of each reservoir is
fixed at 1 mL. Be sure to mix each reservoir
thoroughly when you have finished pipetting. You may do this by
gently swirling the tray in a circular motion. 3) Prepare reservoir
solutions: Pipet the indicated amount of reagent to each of the 24
wells. [(NH4)2SO4] concentration
varies horizontally across a row. The volume of each reservoir is
fixed at 1 mL. Be sure to mix each reservoir
thoroughly when you have finished pipetting. You may do this by
gently swirling the tray in a circular motion.
5) Prepare drops: (a) Place a row of 6 coverslips on the top of
the tray lid, at the edge of the lid. (b)With the P-2 pipet, transfer
3 uL of the proteinase K solution onto the middle
of each cover slip. Use a steady hand to keep the drop in the form of
a nice round bead. The coverslips have been siliconized in order to enhance
bead formation.
 How to avoid blowing
bubbles in the drop: Normally, you expel liquids from the Pipetman by pushing the plunger to the second
stop. The distance between the first stop and the second stop blows
air through the tip which ensures that all the liquid is
expelled. Unfortunately, this feature also blows bubbles in the drop
you are forming on the coverslip (Bubbles mean troubles). You can avoid
blowing bubbles by pushing the plunger to the first stop only.
If you do get a bubble in your drop, you may remove the bubble by holding
the pipet vertically and just touching the top of the drop. How to avoid blowing
bubbles in the drop: Normally, you expel liquids from the Pipetman by pushing the plunger to the second
stop. The distance between the first stop and the second stop blows
air through the tip which ensures that all the liquid is
expelled. Unfortunately, this feature also blows bubbles in the drop
you are forming on the coverslip (Bubbles mean troubles). You can avoid
blowing bubbles by pushing the plunger to the first stop only.
If you do get a bubble in your drop, you may remove the bubble by holding
the pipet vertically and just touching the top of the drop.
 (c)
Pipet 1 uL of reservoir A1 onto the first protein
drop. The protein and reservoir will mix by convection; there is no need to
mix with the Pipetman. Mixing just
increases the likelihood of smearing the drop. (d) Add reservoir A2
to the second drop, etc. (e) Invert the coverslips onto their
respective reservoirs. If you haven't done this before, first try
inverting a coverslip carrying a drop of water. Keep the
coverslip perpendicular to the acceleration, and banked a little. Be
careful to align the coverslips to the edges of the plastic tray. Gently
press the coverslips onto the grease seals around the edges using the point
of a pipet tip. It is best not to use your fingers because they get
greasy and smear the glass, obscuring visibility of the crystals. Don't
press directly in the center of the cover slip. (f) Repeat steps a-e for
rows B, C, and D, but change the volume of ProK
and reservoir according to the instructions on the handout
from Genesis Falcon. 6) Label your tray with your names, the date, and the
protein name using a Sharpie. Crystals
will appear in a few hours and reach full size overnight. (c)
Pipet 1 uL of reservoir A1 onto the first protein
drop. The protein and reservoir will mix by convection; there is no need to
mix with the Pipetman. Mixing just
increases the likelihood of smearing the drop. (d) Add reservoir A2
to the second drop, etc. (e) Invert the coverslips onto their
respective reservoirs. If you haven't done this before, first try
inverting a coverslip carrying a drop of water. Keep the
coverslip perpendicular to the acceleration, and banked a little. Be
careful to align the coverslips to the edges of the plastic tray. Gently
press the coverslips onto the grease seals around the edges using the point
of a pipet tip. It is best not to use your fingers because they get
greasy and smear the glass, obscuring visibility of the crystals. Don't
press directly in the center of the cover slip. (f) Repeat steps a-e for
rows B, C, and D, but change the volume of ProK
and reservoir according to the instructions on the handout
from Genesis Falcon. 6) Label your tray with your names, the date, and the
protein name using a Sharpie. Crystals
will appear in a few hours and reach full size overnight.
7) Next week: One group will collect data on the native crystal.
Another group will collect data on the PMSF complex. All others will
collect data on potential heavy atom derivatives.
Interested students are invited to soak their crystals
in heavy atom solution in the X-ray lab (BH124) in preparation for data
collection and phasing. The experiment takes about 20 minutes. You
can make an appointment with Mike through the google spreadsheet (link
provided at the end of the Crystallization Lab).
|
Part Two:
Preparation
of Heavy Atom Derivatives
|
Objective: Based on the results of the native
gel band shift assay select a heavy atom and soak it into crystals for
derivatization.
Procedures:
1) Look for a band shift in one of the lanes containing a heavy atom.
If the heavy atom complex is cationic, then the protein will have a higher 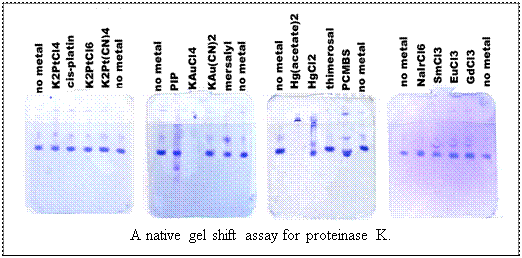 mobility
in the gel (remember, the gel is run with reverse polarity). Lanes in
which no protein appears are the result of aggregation caused by the heavy
atom, and should not be considered further for derivatization. mobility
in the gel (remember, the gel is run with reverse polarity). Lanes in
which no protein appears are the result of aggregation caused by the heavy
atom, and should not be considered further for derivatization.
2) Select the heavy atom which you are most confident caused a shift.
3) Prepare a 1:5 dilution of the selected heavy atom.
4) Select two drops with nice crystals from the tray of proteinase K
crystals that you grew.
5) Add 1 uL of diluted heavy
atom to the selected drops. Dispose of pipet tips in designated waste
containers.
|